Botanical estrogen (BE) dietary supplements are consumed by women as substitutes for loss of endogenous estrogens at menopause. To examine the roles of estrogen receptor α (ERα) and aryl hydrocarbon receptor (AhR) and their crosstalk in the actions of BEs, we studied gene regulation and proliferation responses to four widely used BEs, genistein, daidzein, and S-equol from soy, and liquiritigen from licorice root in breast cancer and liver cells. BEs and estradiol (E2), acting through ERα, stimulated proliferation, ERα chromatin binding and target-gene expression. BEs but not E2, acting through AhR, bound to xenobiotic response element-containing chromatin sites and enhanced AhR target-gene expression (CYP1A1, CYP1B1). While E2 and TCDD acted quite selectively through their respective receptors, BEs acted via both receptors, with their AhR activity moderated by negative crosstalk through ERα. Both ERα and AhR should be considered as mediators of the biology and pharmacology of BEs. Link
Category Archives: Botanical Estrogens
Estrogen and Microbiota Crosstalk: Should We Pay Attention?
Recent advances have suggested that steroid hormones such as estrogens, and gut microbiota might synergize to influence obesity, diabetes, and cancer. We discuss recent knowledge of the interactions between estrogens and gut microbiota, and new insights that might offer new approaches to influence this crosstalk and improve metabolic outcomes. Link
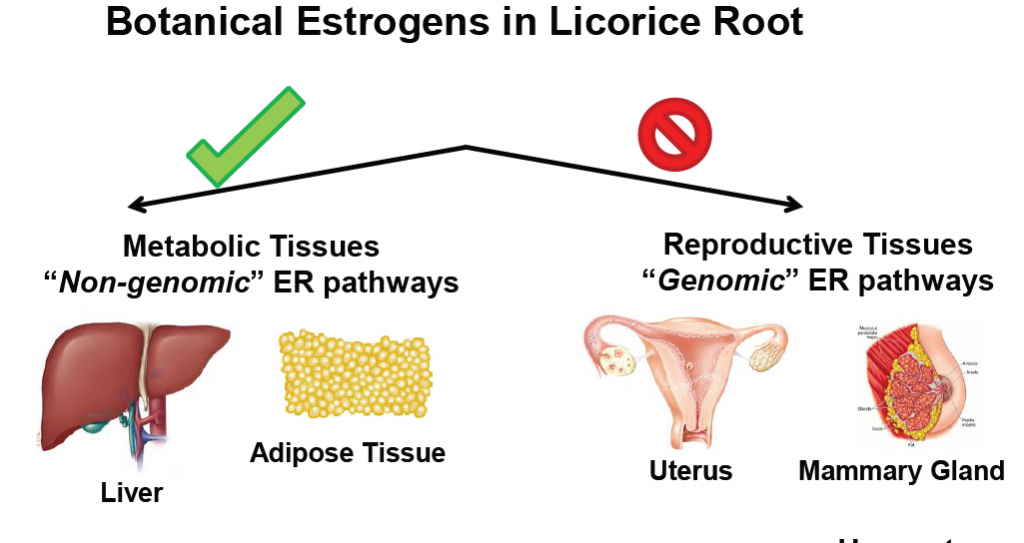
Dietary licorice root supplementation reduces diet-induced weight gain, lipid deposition, and hepatic steatosis in ovariectomized mice without stimulating reproductive tissues and mammary gland
Transcriptomic analysis identifies gene networks regulated by estrogen receptor α (ERα) and ERβ that control distinct effects of different botanical estrogens. Gong P, Madak-Erdogan Z, Li J1, Cheng J1, Greenlief CM1, Helferich WG, Katzenellenbogen JA, Katzenellenbogen B. Nucl Recept Signal. 2014 Sep 12;12S
The estrogen receptors (ERs) ERα and ERβ mediate the actions of endogenous estrogens as well as those of botanical estrogens (BEs) present in plants. BEs are ingested in the diet and also widely consumed by postmenopausal women as dietary supplements, often as a substitute for the loss of endogenous estrogens at menopause. However, their activities and efficacies, and similarities and differences in gene expression programs with respect to endogenous estrogens such as estradiol (E2) are not fully understood. Because gene expression patterns underlie and control the broad physiological effects of estrogens, we have investigated and compared the gene networks that are regulated by different BEs and by E2. Our aim was to determine if the soy and licorice BEs control similar or different gene expression programs and to compare their gene regulations with that of E2. Gene expression was examined by RNA-Seq in human breast cancer (MCF7) cells treated with control vehicle, BE or E2. These cells contained three different complements of ERs, ERα only, ERα+ERβ, or ERβ only, reflecting the different ratios of these two receptors in different human breast cancers and in different estrogen target cells. Using principal component, hierarchical clustering, and gene ontology and interactome analyses, we found that BEs regulated many of the same genes as did E2. The genes regulated by each BE, however, were somewhat different from one another, with some genes being regulated uniquely by each compound. The overlap with E2 in regulated genes was greatest for the soy isoflavones genistein and S-equol, while the greatest difference from E2 in gene expression pattern was observed for the licorice root BE liquiritigenin. The gene expression pattern of each ligand depended greatly on the cell background of ERs present. Despite similarities in gene expression pattern with E2, the BEs were generally less stimulatory of genes promoting proliferation and were more pro-apoptotic in their gene regulations than E2. The distinctive patterns of gene regulation by the individual BEs and E2 may underlie differences in the activities of these soy and licorice-derived BEs in estrogen target cells containing different levels of the two ERs. PMID:25363786
Mechanism Enforcing the Estrogen Receptor Beta-Selectivity of Botanical Estrogens, Jiang, Y., Gong, P., Madak-Erdogan, Z., Martin, T., Jeyakumar, M., Carlson, K., Khan, I., Smillie, T.J., Chittiboyina, A.G., Rotte S.C.K., Helferich, W.G., Katzenellenbogen, J.A., Katzenellenbogen, B.S., FASEB Journal 2013 Nov;27(11):4406-18,
Because little is known about the actions of botanical estrogens (BEs), widely consumed by menopausal women, we investigated the mechanistic and cellular activities of some major BEs. We examined the interactions of genistein, daidzein, equol, and liquiritigenin with estrogen receptors ERα and ERβ, with key coregulators (SRC3 and RIP140) and chromatin binding sites, and the regulation of gene expression and proliferation in MCF-7 breast cancer cells containing ERα and/or ERβ. Unlike the endogenous estrogen, estradiol (E2), BEs preferentially bind to ERβ, but their ERβ-potency selectivity in gene stimulation (340- to 830-fold vs. E2) is enhanced at several levels (coregulator recruitment, chromatin binding); nevertheless, at high (0.1 or 1 μM) concentrations, BEs also fully activate ERα. Because ERα drives breast cancer cell proliferation and ERβ dampens this, the relative levels of these two ERs in target cells and the BE dose greatly affect gene expression and proliferative response and will be crucial determinants of the potential benefits vs. risks of BEs. Our findings reveal key and novel mechanistic differences in the estrogenic activities of BEs vs. E2, with BEs displaying patterns of activity distinctly different from those seen with E2 and provide valuable information to inform future studies PMID: 23882126
Integrative Genomics of Gene and Metabolic Regulation by Estrogen Receptors α and β and Coregulators, Madak-Erdogan Z, Charn T.H, Jiang Y, Liu E.T., Katzenellenbogen J.A., Katzenellenbogen B.S.Molecular Systems Biology 2013 Jun 18;9:676,
The closely related transcription factors (TFs), estrogen receptors ERα and ERβ, regulate divergent gene expression programs and proliferative outcomes in breast cancer. Utilizing breast cancer cells with ERα, ERβ, or both receptors as a model system to define the basis for differing response specification by related TFs, we show that these TFs and their key coregulators, SRC3 and RIP140, generate overlapping as well as unique chromatin-binding and transcription-regulating modules. Cistrome and transcriptome analyses and the use of clustering algorithms delineated 11 clusters representing different chromatin-bound receptor and coregulator assemblies that could be functionally associated through enrichment analysis with distinct patterns of gene regulation and preferential coregulator usage, RIP140 with ERβ and SRC3 with ERα. The receptors modified each other’s transcriptional effect, and ERβ countered the proliferative drive of ERα through several novel mechanisms associated with specific binding-site clusters. Our findings delineate distinct TF-coregulator assemblies that function as control nodes, specifying precise patterns of gene regulation, proliferation, and metabolism, as exemplified by two of the most important nuclear hormone receptors in human breast cancer. PMID: 23774759